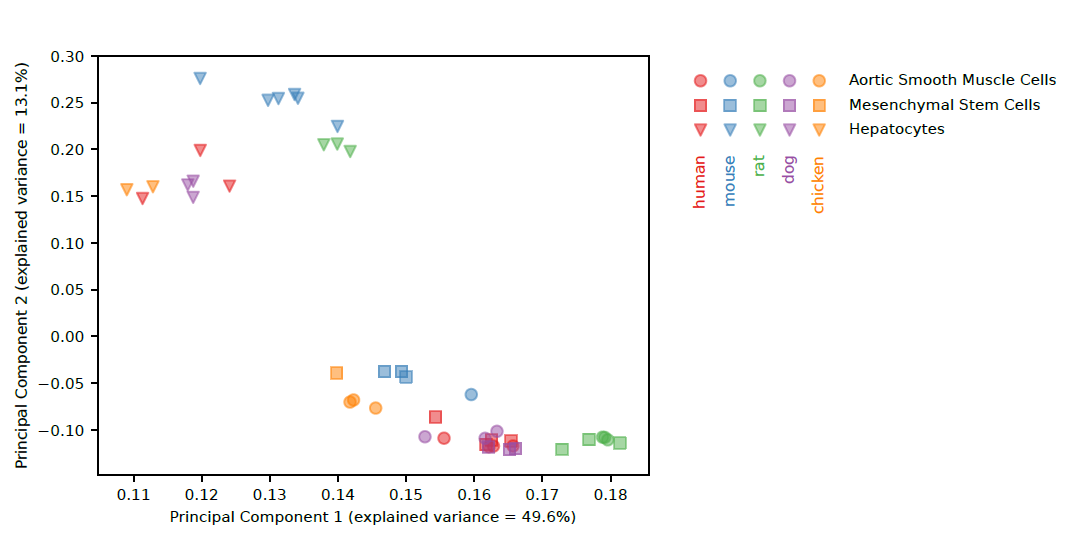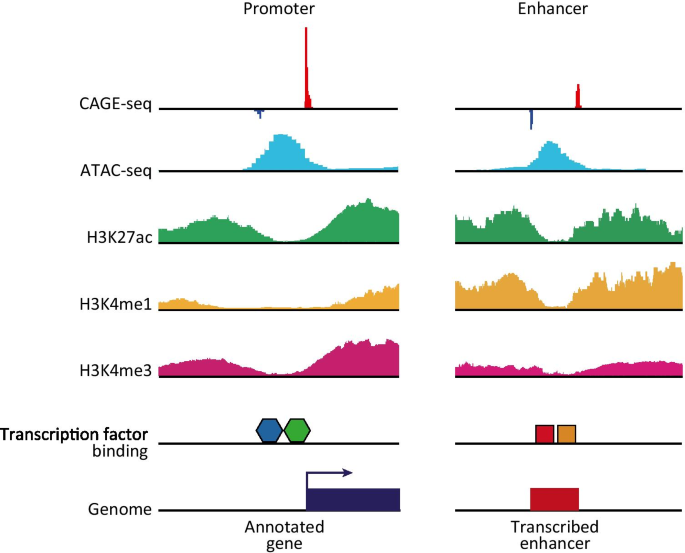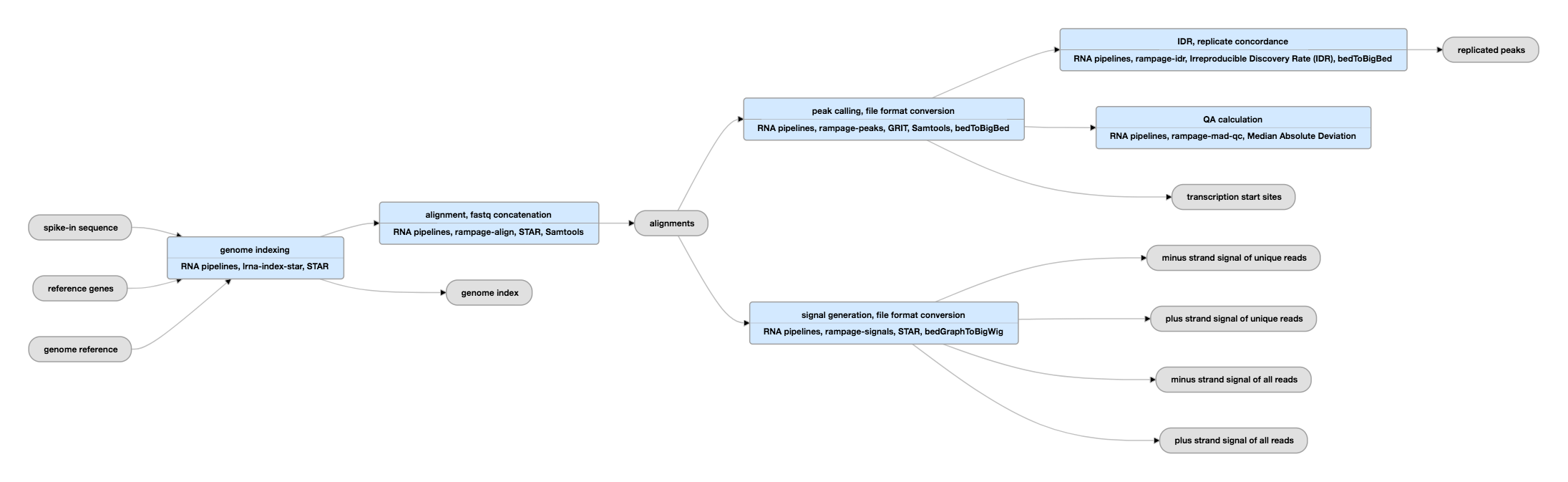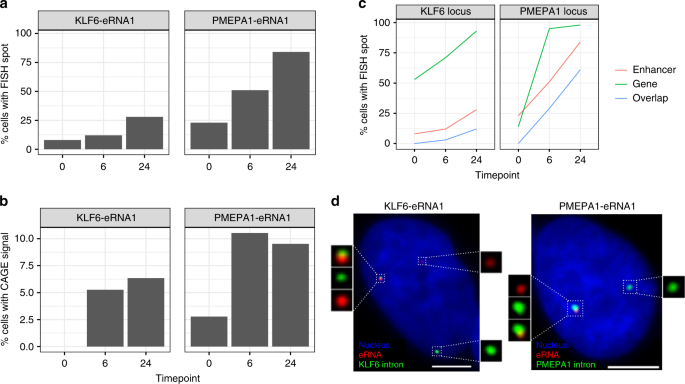
C1 CAGE detects transcription start sites and enhancer activity at single-cell resolution | Nature Communications
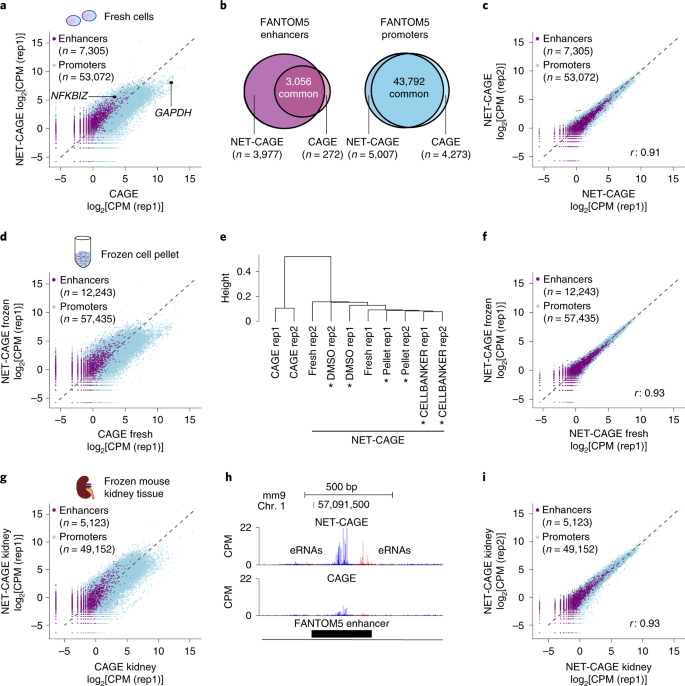
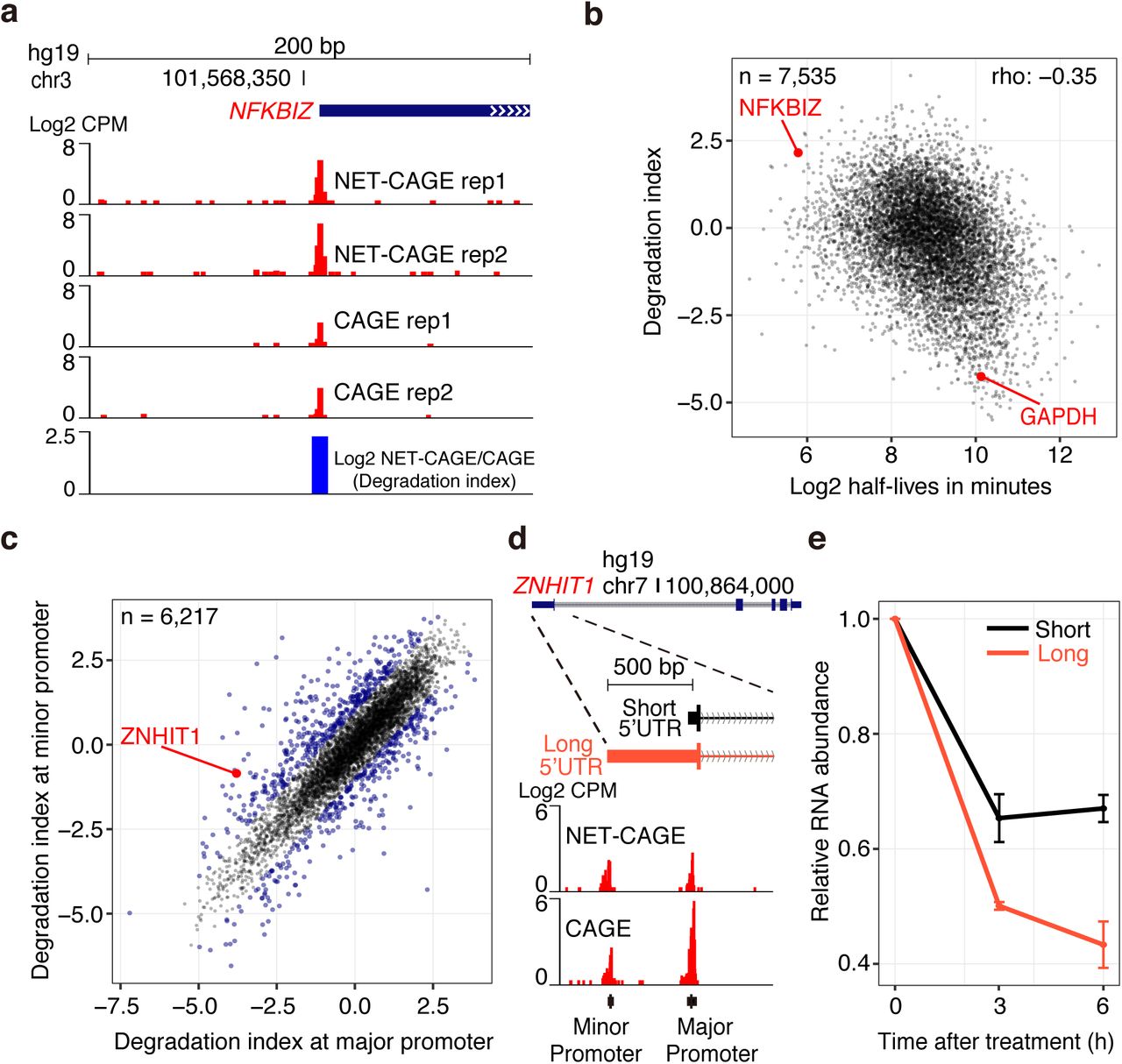
NET-CAGE characterizes the dynamics and topology of human transcribed cis-regulatory elements | Nature Genetics

CAGE-seq characterization of alternative promoter usage and full-length... | Download Scientific Diagram
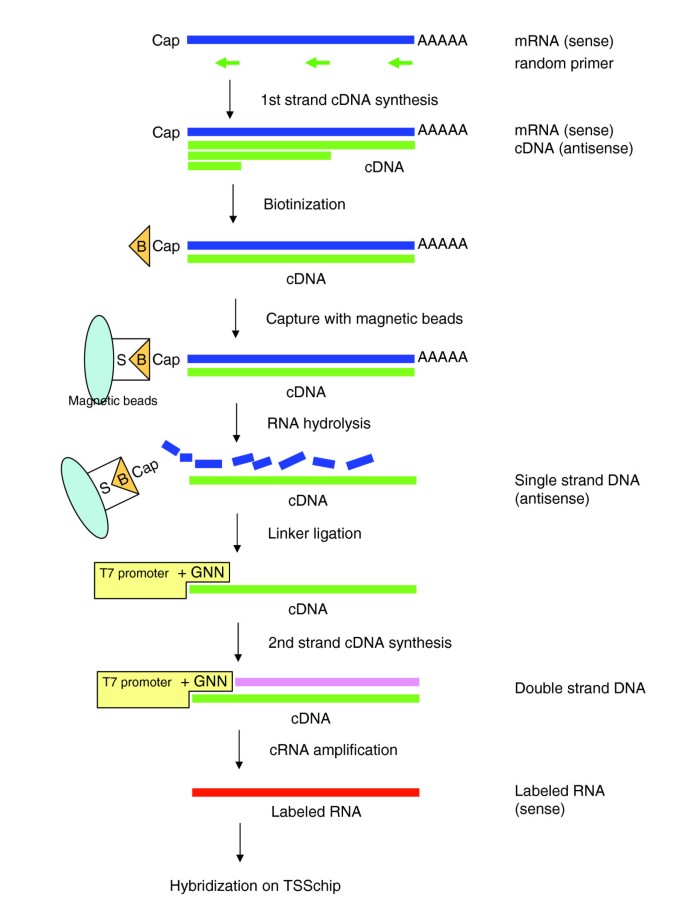
CAGE-TSSchip: promoter-based expression profiling using the 5'-leading label of capped transcripts | Genome Biology | Full Text
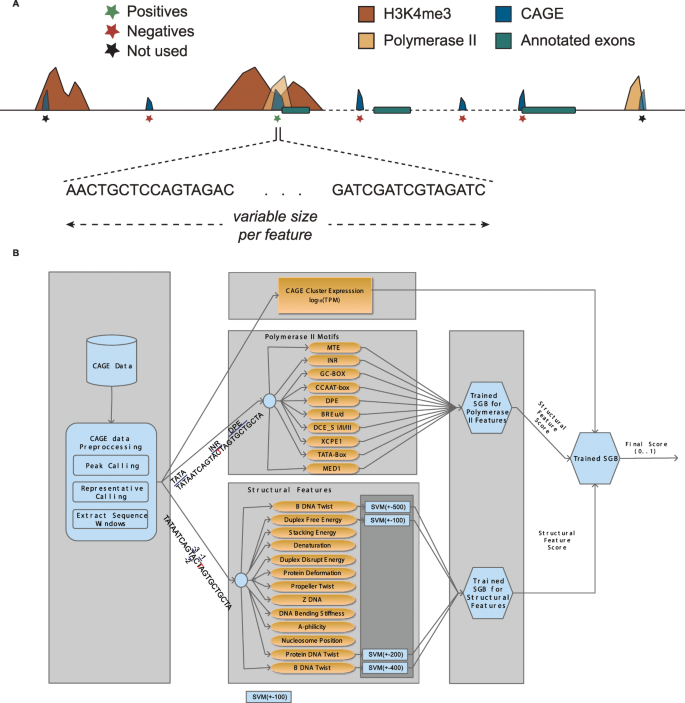
Solving the transcription start site identification problem with ADAPT-CAGE: a Machine Learning algorithm for the analysis of CAGE data | Scientific Reports

Cap analysis of gene expression reveals alternative promoter usage in a rat model of hypertension | Life Science Alliance

Solving the transcription start site identification problem with ADAPT-CAGE: a Machine Learning algorithm for analysis of CAGE data | bioRxiv

Characterization of Arabidopsis thaliana promoter bidirectionality and antisense RNAs by depletion of nuclear RNA decay enzymes | bioRxiv
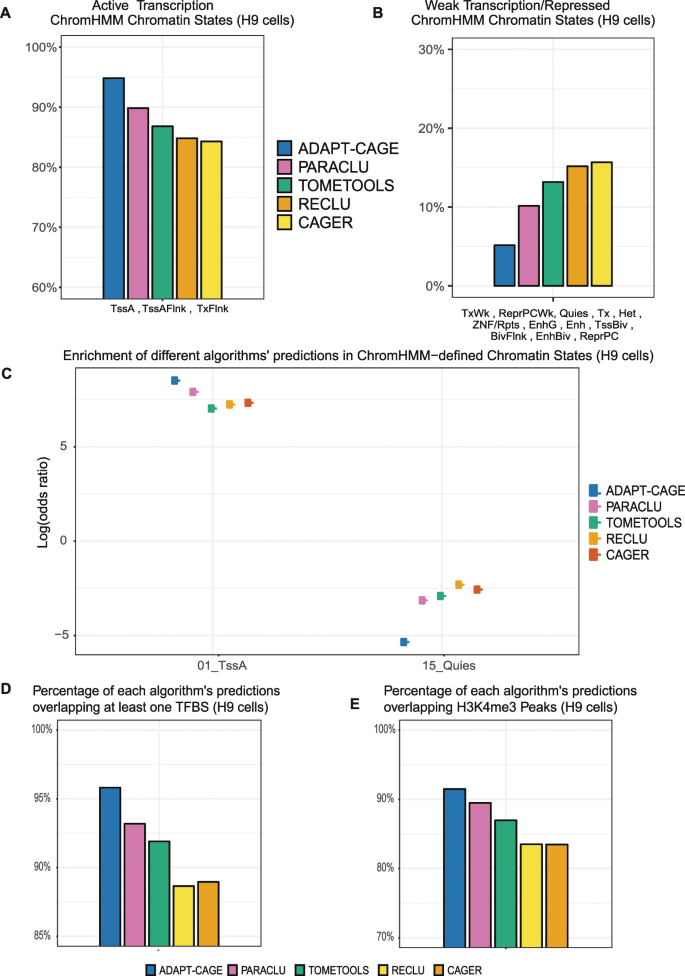
Solving the transcription start site identification problem with ADAPT-CAGE: a Machine Learning algorithm for the analysis of CAGE data | Scientific Reports
PLOS Computational Biology: Influence of Rotational Nucleosome Positioning on Transcription Start Site Selection in Animal Promoters

Deep Cap Analysis of Gene Expression (CAGE): Genome-Wide Identification of Promoters, Quantification of Their Activity, and Transcriptional Network Inference | SpringerLink
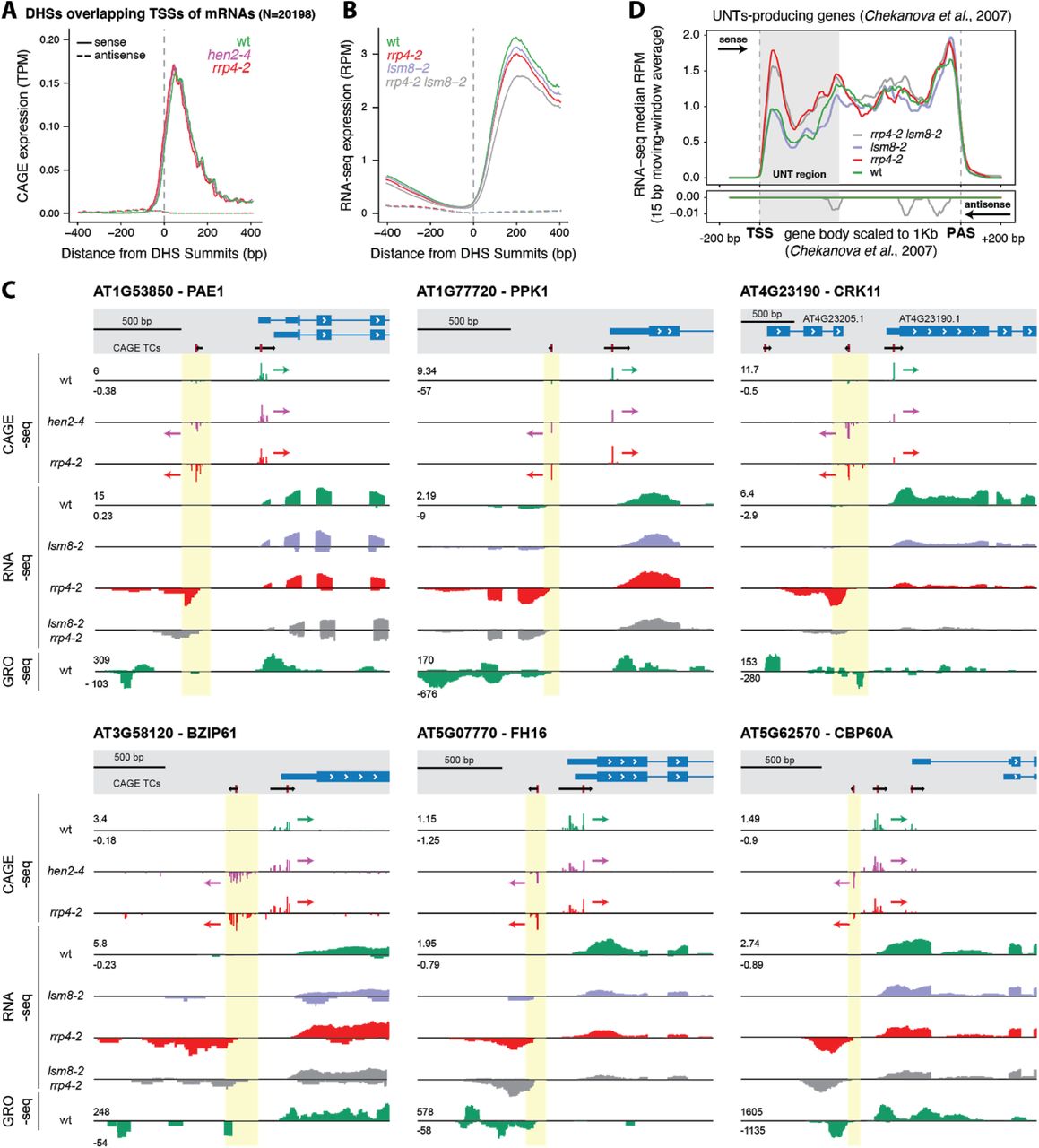
Characterization of Arabidopsis thaliana promoter bidirectionality and antisense RNAs by depletion of nuclear RNA decay enzymes | bioRxiv

Deep cap analysis gene expression (CAGE): genome-wide identification of promoters, quantification of their expression, and network inference | BioTechniques
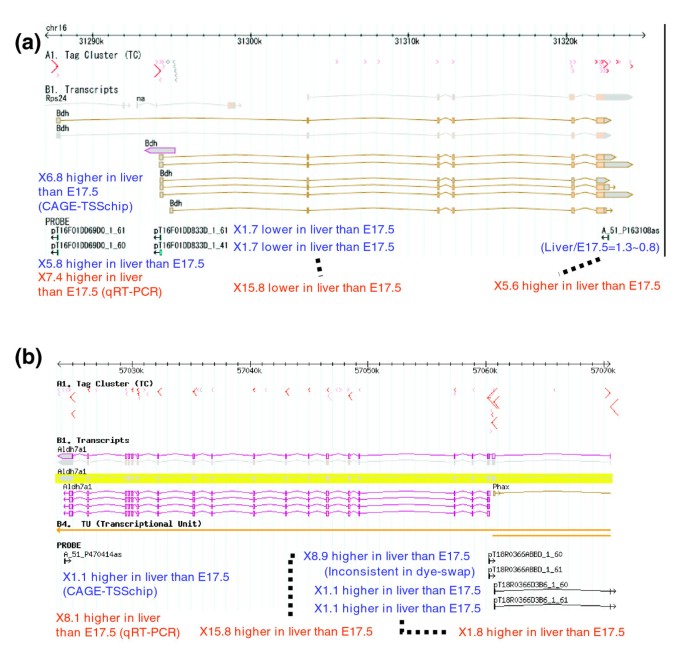
CAGE-TSSchip: promoter-based expression profiling using the 5'-leading label of capped transcripts | Genome Biology | Full Text